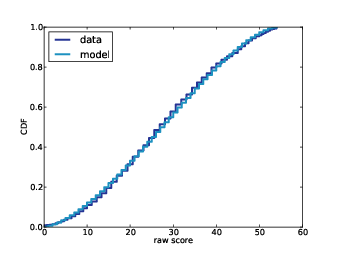
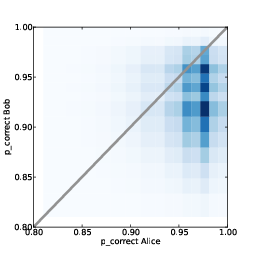
In Part One I presented the simplest version of the algorithm, which I think is easy to understand, but slow. In Think Bayes I present some ways to optimize it. In Part Two I apply the algorithm to real data and generate predictive distributions. Now in Part Three, as promised, I publish the predictions the algorithm generates. In Part Four I will compare the predictions to actual data.
Background: Belly Button Biodiversity 2.0 (BBB2) is a nation-wide citizen science project with the goal of identifying bacterial species that can be found in human navels (http://bbdata.yourwildlife.org).
Transparent science
In an effort to explore the limits of transparent science, I have started publishing my research in this blog as I go along. This past summer I wrote a series of articles exploring the relationship between Internet use and religious disaffiliation. This "publish as you go" model should help keep researchers honest. Among other things, it might mitigate publication bias due to the "file drawer effect." And if the data and code are published along with the results, that should help make experiments more reproducible.Toward that end, I will now subject myself to public humiliation by generating a set of predictions using my almost-entirely-unvalidated solution to the Unseen Species problem. In the next installment I will publish the correct answers and score my predictions. Here are the details:
1) I am working with data from the Belly Button Biodiversity project; this data was used in a paper published in PLOS ONE and made available on the web pages of the journal and the researchers. The data consists of rDNA "reads" from 60 subjects. In order to facilitate comparisons between subjects, the researchers chose subjects with at least 400 reads, and for each subject they chose a random subset of 400 reads. The data for the other reads was not published.
2) For each subject, I know the results of the 400 selected reads, and the total number of reads. I will use my algorithm to generate a "prediction" for each subject, which is the number of additional species in the complete dataset.
3) Specifically, for each subject I will generate 9 posterior credible intervals (CIs) for the number of additional species: a 10% CI, a 20% CI, and so on up to a 90% CI.
4) To validate my predictions, I will count the number of CIs that contain the actual, correct value. Ideally, 10% of the correct values should fall in the 10% CIs, 20% should fall in the 20% CIs, and so on. Since the predictions and actual values are integers, a value that hits one end of a predicted CI counts as a half-hit.
Predictions
And now, without further ado, here are my predictions. The columns labelled 10, 20, etc. are 10% credible intervals, 20% CIs, and so on.| Code | # reads | # species | 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| B1234 | 1392 | 48 | (4, 4) | (4, 5) | (3, 5) | (3, 5) | (3, 6) | (2, 6) | (2, 7) | (1, 7) | (1, 9) |
| B1235 | 2452 | 69 | (11, 12) | (10, 12) | (10, 13) | (9, 13) | (8, 14) | (8, 15) | (7, 16) | (6, 17) | (5, 19) |
| B1236 | 2964 | 45 | (4, 5) | (4, 5) | (4, 6) | (4, 6) | (3, 7) | (3, 7) | (3, 8) | (2, 9) | (1, 10) |
| B1237 | 3090 | 62 | (9, 10) | (9, 11) | (8, 11) | (8, 11) | (7, 12) | (7, 12) | (6, 13) | (5, 14) | (4, 16) |
| B1242 | 3056 | 61 | (9, 9) | (8, 10) | (8, 10) | (7, 11) | (7, 11) | (6, 12) | (6, 14) | (5, 15) | (5, 16) |
| B1243 | 1518 | 71 | (10, 11) | (10, 12) | (9, 12) | (8, 13) | (8, 13) | (8, 14) | (7, 15) | (6, 16) | (5, 17) |
| B1246 | 4230 | 91 | (23, 24) | (22, 25) | (21, 26) | (21, 27) | (19, 28) | (18, 29) | (17, 30) | (16, 33) | (14, 35) |
| B1253 | 1928 | 86 | (16, 17) | (15, 18) | (14, 18) | (14, 20) | (13, 20) | (13, 21) | (12, 23) | (11, 24) | (10, 26) |
| B1254 | 918 | 58 | (5, 5) | (4, 6) | (4, 6) | (3, 6) | (3, 7) | (3, 7) | (2, 8) | (2, 9) | (1, 10) |
| B1258 | 1350 | 87 | (15, 16) | (14, 17) | (14, 17) | (13, 18) | (12, 19) | (11, 19) | (11, 20) | (10, 21) | (8, 24) |
| B1259 | 1002 | 80 | (10, 11) | (10, 12) | (10, 12) | (9, 13) | (9, 14) | (8, 14) | (7, 15) | (6, 16) | (6, 18) |
| B1260 | 1944 | 96 | (22, 23) | (21, 24) | (20, 25) | (19, 25) | (19, 26) | (18, 27) | (17, 29) | (15, 30) | (14, 32) |
| B1264 | 1122 | 83 | (12, 13) | (12, 14) | (11, 14) | (10, 15) | (10, 15) | (9, 16) | (8, 17) | (7, 18) | (6, 20) |
| B1265 | 2928 | 85 | (18, 19) | (17, 20) | (16, 21) | (16, 22) | (15, 23) | (14, 24) | (13, 25) | (12, 26) | (11, 28) |
| B1273 | 2980 | 61 | (9, 9) | (8, 10) | (8, 10) | (7, 11) | (7, 12) | (6, 12) | (6, 13) | (5, 14) | (4, 16) |
| B1275 | 1672 | 85 | (16, 17) | (15, 18) | (15, 19) | (14, 19) | (13, 20) | (13, 21) | (12, 22) | (11, 24) | (9, 25) |
| B1278 | 1242 | 47 | (4, 4) | (3, 4) | (3, 5) | (3, 5) | (2, 6) | (2, 6) | (2, 6) | (2, 7) | (1, 8) |
| B1280 | 1772 | 46 | (4, 4) | (4, 5) | (3, 5) | (3, 5) | (3, 6) | (2, 6) | (2, 7) | (2, 8) | (1, 9) |
| B1282 | 1132 | 67 | (8, 9) | (7, 9) | (7, 10) | (6, 10) | (6, 11) | (6, 11) | (5, 12) | (5, 13) | (3, 15) |
| B1283 | 1414 | 67 | (8, 9) | (8, 10) | (7, 10) | (7, 11) | (7, 11) | (6, 12) | (5, 13) | (4, 14) | (3, 16) |
| B1284 | 1158 | 91 | (15, 16) | (14, 17) | (14, 17) | (13, 18) | (13, 19) | (12, 19) | (12, 20) | (10, 21) | (9, 23) |
| B1285 | 2340 | 55 | (7, 7) | (6, 8) | (6, 8) | (5, 8) | (5, 9) | (4, 9) | (4, 10) | (3, 12) | (2, 13) |
| B1286 | 1728 | 66 | (9, 10) | (9, 11) | (8, 11) | (8, 12) | (8, 12) | (7, 13) | (6, 14) | (6, 14) | (4, 16) |
| B1288 | 1280 | 107 | (23, 24) | (22, 25) | (21, 25) | (21, 26) | (20, 27) | (19, 27) | (18, 29) | (17, 31) | (15, 32) |
| B1289 | 2054 | 103 | (26, 27) | (25, 28) | (24, 29) | (23, 30) | (23, 30) | (22, 32) | (21, 33) | (20, 34) | (17, 36) |
| B1291 | 1248 | 94 | (17, 18) | (16, 19) | (16, 20) | (15, 20) | (15, 21) | (13, 22) | (13, 23) | (12, 25) | (10, 27) |
| B1292 | 1864 | 82 | (15, 16) | (14, 16) | (13, 17) | (13, 18) | (13, 19) | (12, 20) | (11, 21) | (10, 22) | (9, 24) |
| B1293 | 1904 | 76 | (13, 14) | (12, 14) | (12, 15) | (11, 16) | (11, 16) | (10, 17) | (9, 18) | (8, 19) | (7, 22) |
| B1294 | 1784 | 78 | (14, 15) | (13, 16) | (12, 16) | (12, 17) | (11, 18) | (11, 19) | (10, 19) | (9, 20) | (8, 23) |
| B1295 | 1408 | 70 | (10, 10) | (9, 11) | (9, 12) | (8, 12) | (8, 12) | (7, 13) | (7, 14) | (6, 15) | (4, 17) |
| B1296 | 2034 | 55 | (7, 7) | (6, 8) | (6, 8) | (6, 8) | (5, 9) | (4, 9) | (4, 10) | (4, 11) | (3, 12) |
| B1298 | 1478 | 72 | (10, 11) | (9, 12) | (9, 12) | (9, 13) | (8, 13) | (8, 14) | (7, 15) | (6, 16) | (5, 18) |
| B1308 | 1160 | 58 | (6, 6) | (5, 7) | (5, 7) | (5, 7) | (4, 8) | (4, 8) | (3, 9) | (3, 10) | (2, 11) |
| B1310 | 1066 | 80 | (11, 12) | (11, 13) | (10, 13) | (9, 14) | (9, 15) | (8, 15) | (7, 16) | (7, 17) | (5, 19) |
| B1374 | 2364 | 48 | (5, 5) | (4, 6) | (4, 6) | (4, 6) | (3, 7) | (3, 7) | (3, 8) | (2, 9) | (2, 10) |
| B940 | 2874 | 93 | (22, 24) | (21, 25) | (21, 25) | (20, 26) | (19, 27) | (19, 28) | (18, 30) | (16, 32) | (14, 33) |
| B941 | 2154 | 48 | (5, 6) | (5, 6) | (4, 6) | (4, 7) | (4, 7) | (3, 7) | (3, 8) | (2, 9) | (2, 11) |
| B944 | 954 | 52 | (4, 4) | (4, 5) | (3, 5) | (3, 5) | (3, 6) | (2, 6) | (2, 6) | (2, 7) | (1, 9) |
| B945 | 2390 | 67 | (10, 11) | (10, 12) | (9, 12) | (9, 13) | (8, 13) | (8, 14) | (7, 15) | (7, 16) | (5, 17) |
| B946 | 5012 | 85 | (20, 21) | (19, 22) | (19, 23) | (18, 24) | (18, 24) | (17, 26) | (16, 27) | (15, 28) | (12, 31) |
| B947 | 3356 | 62 | (10, 11) | (9, 11) | (9, 12) | (8, 12) | (7, 13) | (7, 14) | (6, 14) | (5, 15) | (5, 17) |
| B948 | 2384 | 80 | (16, 17) | (15, 18) | (14, 18) | (14, 19) | (13, 20) | (12, 21) | (11, 22) | (10, 23) | (9, 25) |
| B950 | 1560 | 63 | (8, 9) | (8, 10) | (8, 10) | (7, 10) | (7, 11) | (6, 11) | (5, 12) | (5, 13) | (4, 15) |
| B952 | 1648 | 57 | (7, 7) | (6, 8) | (6, 8) | (6, 8) | (5, 9) | (5, 9) | (4, 10) | (3, 11) | (3, 12) |
| B953 | 1452 | 32 | (2, 2) | (1, 2) | (1, 2) | (1, 3) | (1, 3) | (1, 3) | (0, 3) | (0, 4) | (0, 5) |
| B954 | 1996 | 29 | (2, 2) | (1, 2) | (1, 2) | (1, 2) | (1, 3) | (1, 3) | (0, 3) | (0, 4) | (0, 4) |
| B955 | 1474 | 65 | (8, 9) | (8, 9) | (7, 9) | (7, 10) | (7, 10) | (6, 11) | (5, 12) | (5, 13) | (4, 14) |
| B956 | 1482 | 71 | (10, 11) | (10, 12) | (10, 12) | (9, 13) | (8, 14) | (8, 14) | (7, 15) | (6, 16) | (5, 18) |
| B957 | 2604 | 36 | (3, 3) | (3, 3) | (2, 4) | (2, 4) | (2, 5) | (1, 5) | (1, 6) | (1, 6) | (1, 7) |
| B958 | 2840 | 29 | (2, 2) | (2, 2) | (1, 2) | (1, 3) | (1, 3) | (1, 3) | (1, 4) | (0, 4) | (0, 5) |
| B961 | 1214 | 36 | (2, 3) | (2, 3) | (2, 3) | (2, 4) | (1, 4) | (1, 4) | (1, 5) | (1, 5) | (0, 6) |
| B962 | 1138 | 41 | (3, 3) | (2, 3) | (2, 3) | (2, 4) | (2, 4) | (1, 4) | (1, 5) | (1, 6) | (0, 7) |
| B963 | 1600 | 71 | (10, 12) | (10, 12) | (9, 12) | (9, 13) | (8, 14) | (8, 14) | (7, 15) | (5, 16) | (4, 19) |
| B966 | 1950 | 80 | (15, 16) | (14, 16) | (14, 17) | (13, 17) | (12, 18) | (11, 18) | (11, 20) | (10, 22) | (9, 23) |
| B967 | 1108 | 47 | (3, 4) | (3, 4) | (3, 4) | (2, 5) | (2, 5) | (2, 5) | (2, 6) | (1, 7) | (1, 7) |
| B968 | 2432 | 52 | (6, 7) | (6, 7) | (6, 7) | (5, 8) | (5, 8) | (4, 9) | (3, 9) | (3, 10) | (2, 11) |
| B971 | 1462 | 49 | (4, 5) | (4, 5) | (4, 5) | (3, 6) | (3, 6) | (3, 7) | (2, 7) | (2, 8) | (1, 9) |
| B972 | 1438 | 75 | (11, 12) | (11, 13) | (10, 13) | (10, 14) | (9, 14) | (8, 15) | (8, 16) | (7, 17) | (6, 18) |
| B974 | 5072 | 54 | (7, 8) | (7, 9) | (6, 9) | (6, 9) | (5, 10) | (5, 10) | (4, 11) | (4, 12) | (2, 14) |
| B975 | 1542 | 63 | (7, 8) | (7, 9) | (6, 9) | (6, 9) | (6, 10) | (5, 11) | (5, 11) | (4, 12) | (3, 14) |
Reading the last row, subject B975 yielded 1542 reads; in a random subset of 400 reads, there were 63 different species (or more precisely, OTUs). My algorithm predicts that if we look at all 1542 reads, the number of additional species we'll find is between 3 and 14, with 90% confidence.
I have to say that this table fills me with dread. The intervals seem quite small, which is to say that the algorithm is more confident than I am. The 90% CIs seem especially narrow to me; it is hard for me to believe that 90% of them will contain the correct values. Well, I guess that's why Karl Popper called them "bold hypotheses". We'll find out soon whether they are bold, or just reckless.
I want to thank Rob Dunn at BBB2 for his help with this project. The code and data I used to generate these results are available from this SVN repository.
EDIT 2-22-13: I ran the predictions again with more simulations. The results are not substantially different. I still haven't looked at the answers.